The samples collected and analyzed as part of FishPopTrace are stored in a series of databases. Access to the scientific raw data and analysis is currently restricted to FishPopTrace partners. A website “Member Area” acts as an online secure file store, allowing FishPopTrace partners to collaborate on documents and to distribute files amongst each other.
Below, a short description of the database structures and the various user interfaces are given. Although the structures described below contain FishPopTrace data pertaining initially to a limited number of samples (7,500) spread over 4 commercial species, they are designed to be expanded and continually built on, to work towards building a full genetic characterization of the marine environment.
The characteristics of the fish specimen collected are stored in a central “sampling database”. This information describes the specimen age, length, weight, reproductive characteristics, location and depth of catch, storage characteristics, etc.
Results and raw data for each of the genetic analysis techniques applied to these samples are stored in a series of satellite databases representing each of the techniques employed by FishPopTrace.
This cross referencing approach allows results arising from SNPs, otoliths, proteomics, fatty acids and gene expression analyses to be compared for the same sample.
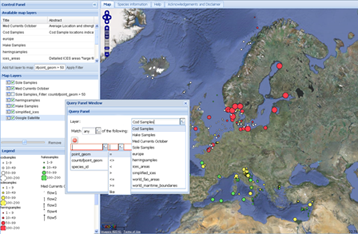
The primary means of information dissemination is through a publicly accessible web-based GeoBrowser.
Information on the samples taken during FishPopTrace, and their genetic characteristics are visualized in an interactive map.
The data visible through this interface is publicly accessible and has a limited degree of access to the genetic analysis information in the databases.
Environmental parameters such as sea surface temperature chlorophyll concentration and ocean currents, along with important international boundaries and economic zones are also available through this interface. This allows possible inferences regarding the effects of environmental parameters on the genetic characteristics of a species to be generated, while also considering what economic or management areas are involved.
A password protected shared file repository allows members of the FishPopTrace consortium save information in a communal file store.
This is a useful function for the creation of joint reports and the storage of consortium specific documents such as templates, presentations, publications and imagery for use in communications.